
Yang Lab | Recording & Decoding Cancer Evolution


Research & Discoveries
We are generally interested in investigating the fundamental principles of tumor evolution and identifying key regulators of tumor progression. Our research is at the interface of cancer biology, technology development, and computational analysis, which combines CRISPR-based molecular recording tools, genetically engineered mouse models (GEMMs), single cell genomics-related algorithm development, and in vivo functional assays.
Major research questions:
Tumor evolution is driven by the acquisition of phenotypic heterogeneity in cancer cells, leading to increased aggressiveness, invasion, immune evasion, therapeutic resistance, and metastasis. However, the molecular mechanisms governing cancer cell state transitions and tumor evolution remain elusive.
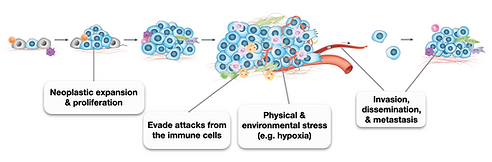
Our lab is focused on addressing critical questions related to tumor evolution, including:
1. How does a single transformed cell expand and develop into an aggressive tumor?
2. What are the evolutionary origins and interrelations of different cell states, and how do they contribute to tumor progression?
3. How do the tumor microenvironment and anti-cancer therapies alter cancer cell states and tumor evolution trajectories?
Models of cancer:
To study tumor progression, we utilize genetically engineered mouse models (GEMMs) of cancer. GEMMs offer the advantage of allowing us to investigate tumor evolution within the context of the native microenvironment and under experimentally defined conditions. These models faithfully model the major steps of tumor evolution from nascent cell transformation to aggressive metastasis, recapitulating human tumor progression both molecularly and histopathologically. Furthermore, our lab is actively developing sophisticated GEMMs to enable precise dissection of tumor progression. In addition to GEMMs, we also leverage organoid models, patient-derived xenografts, and human tumor samples to probe the fundamental mechanisms underlying tumor evolution further. These complementary models contribute to a comprehensive understanding of tumor biology and facilitate the exploration of therapeutic strategies.

Technological innovations:
One of the major challenges in studying tumor evolution is the transient nature of many processes and their embedding within the long history of tumor evolutionary events. To overcome this, our lab has developed and applied a CRISPR-based molecular recorder to track the lineage relationships of cancer cells. This innovative tool leverages Cas9-induced DNA cleavage and subsequent repair to progressively generate heritable insertions and deletions at synthetic DNA target sites engineered into the genomes of living cells. Importantly, these DNA target sites are transcribed into poly-adenylated mRNAs, allowing them to be captured along with all other mRNAs using single-cell RNA sequencing (scRNA-seq). This approach allows one to directly link the current cell state (measured by scRNA-seq) with its past lineage history (captured by the lineage tracer), therefore allowing for quantitative assessment of the types and frequency of cell state transitions in evolving tumors.

Research focus and ongoing efforts:
The main goal of the Yang lab is to investigate the intrinsic and extrinsic mechanisms governing cancer cell state transitions and ultimately develop a quantitative roadmap of tumor evolution.
We have recently developed an autochthonous “KP-Tracer” mouse model, which allows us to continuously monitor the processes by which a single cell harboring oncogenic mutations evolves into an aggressive tumor. In collaboration with Nir Yosef's lab, we have developed a new computational algorithm "Cassiopeia" for building high-resolution tumor phylogenies. This offers a significant advance in tumor evolution modeling by enabling quantitative inference of fitness landscapes, cellular plasticity, evolutionary paths of primary tumors and metastases, and the role of any gene of interest in altering all these facets of tumor development.
Building upon these cutting-edge technologies, our lab aims to develop a comprehensive understanding of tumor evolution by integrating multiple data modalities. We will dissect the cell-intrinsic and extrinsic mechanisms that govern cancer cell state transitions, and identify the regulatory gene networks that drive cancer evolution. Furthermore, our lab is committed to expanding the molecular recording toolkits, enabling more precise and detailed reconstruction of tumor life histories. Through these collaborative efforts, we strive to reconstruct the entire trajectory of tumor development, capturing the process from a single transformed cell to a complex and aggressive tumor population, with unprecedented scale and resolution. Ultimately, this holistic approach will pave the way for the development of predictive models of tumor evolution, advancing our ability to predict and potentially intervene in cancer progression.

